activator proteins bind to what type of dna sequence
A transcriptional activator is a protein (transcription factor) that increases transcription of a gene or set of genes.[1] Activators are considered to have positive control over gene expression, as they part to promote cistron transcription and, in some cases, are required for the transcription of genes to occur.[i] [2] [iii] [four] Virtually activators are Dna-binding proteins that bind to enhancers or promoter-proximal elements.[1] The DNA site bound by the activator is referred to equally an "activator-bounden site".[3] The part of the activator that makes protein–poly peptide interactions with the general transcription machinery is referred to as an "activating region" or "activation domain".[1]
Most activators function by binding sequence-specifically to a regulatory Dna site located about a promoter and making protein–protein interactions with the full general transcription mechanism (RNA polymerase and general transcription factors), thereby facilitating the binding of the general transcription mechanism to the promoter.[1] [2] [3] [4] Other activators help promote gene transcription by triggering RNA polymerase to release from the promoter and proceed forth the Dna.[2] At times, RNA polymerase can break before long afterward leaving the promoter; activators too function to allow these "stalled" RNA polymerases to continue transcription.[ane] [2]
The activity of activators tin can be regulated. Some activators have an allosteric site and can only function when a certain molecule binds to this site, essentially turning the activator on.[iv] Postal service-translational modifications to activators can also regulate activity, increasing or decreasing activity depending on the blazon of modification and activator beingness modified.[1]
In some cells, normally eukaryotes, multiple activators can bind to the binding-site; these activators tend to bind cooperatively and interact synergistically.[1] [two]
Structure [edit]
Activator proteins consist of 2 chief domains: a DNA-binding domain that binds to a DNA sequence specific to the activator, and an activation domain that functions to increase cistron transcription by interacting with other molecules.[1] Activator Deoxyribonucleic acid-binding domains come in a multifariousness of conformations, including the helix-turn-helix, zinc finger, and leucine attachment amidst others.[1] [2] [3] These Deoxyribonucleic acid-binding domains are specific to a certain DNA sequence, allowing activators to turn on only sure genes.[1] [ii] [3] Activation domains also come in a variety of types that are categorized based on the domain's amino acid sequence, including alanine-rich, glutamine-rich, and acidic domains.[i] These domains are not as specific, and tend to interact with a diverseness of target molecules.[i]
Activators can besides accept allosteric sites that are responsible for turning the activators themselves on and off.[4]
Mechanism of activity [edit]
Activator binding to regulatory sequences [edit]
Inside the grooves of the DNA double helix, functional groups of the base pairs are exposed.[ii] The sequence of the DNA thus creates a unique pattern of surface features, including areas of possible hydrogen bonding, ionic bonding, as well as hydrophobic interactions.[2] Activators too have unique sequences of amino acids with side chains that are able to interact with the functional groups in Deoxyribonucleic acid.[2] [iii] Thus, the pattern of amino acrid side chains making upwards an activator protein will be complementary to the surface features of the specific DNA regulatory sequence information technology was designed to bind to.[one] [two] [three] The complementary interactions between the amino acids of the activator protein and the functional groups of the Dna create an "exact-fit" specificity between the activator and its regulatory Deoxyribonucleic acid sequence.[2]
Near activators bind to the major grooves of the double helix, every bit these areas tend to be wider, but there are some that will bind to the minor grooves.[1] [2] [iii]
Activator-binding sites may be located very close to the promoter or numerous base pairs abroad.[2] [iii] If the regulatory sequence is located far away, the DNA will loop over itself (Dna looping) in order for the bound activator to interact with the transcription machinery at the promoter site.[two] [3]
In prokaryotes, multiple genes can be transcribed together (operon), and are thus controlled under the same regulatory sequence.[2] In eukaryotes, genes tend to exist transcribed individually, and each gene is controlled by its ain regulatory sequences.[2] Regulatory sequences where activators bind are usually found upstream from the promoter, but they tin also exist plant downstream or even inside introns in eukaryotes.[1] [ii] [iii]
Functions to increase gene transcription [edit]
Binding of the activator to its regulatory sequence promotes gene transcription past enabling RNA polymerase activity.[1] [2] [3] [4] This is done through diverse mechanisms, such every bit recruiting transcription machinery to the promoter and triggering RNA polymerase to continue into elongation.[1] [two] [iii] [4]
Recruitment [edit]
Activator-controlled genes require the bounden of activators to regulatory sites in order to recruit the necessary transcription machinery to the promoter region.[ane] [2] [iii]
Activator interactions with RNA polymerase are mostly direct in prokaryotes and indirect in eukaryotes.[ii] In prokaryotes, activators tend to make contact with the RNA polymerase directly in order to help bind information technology to the promoter.[2] In eukaryotes, activators mostly interact with other proteins, and these proteins will then be the ones to collaborate with the RNA polymerase.[two]
Prokaryotes [edit]
In prokaryotes, genes controlled past activators accept promoters that are unable to strongly bind to RNA polymerase past themselves.[2] [3] Thus, activator proteins aid to promote the binding of the RNA polymerase to the promoter.[2] [iii] This is done through diverse mechanisms. Activators may bend the Deoxyribonucleic acid in society to amend expose the promoter then the RNA polymerase can demark more than effectively.[3] Activators may make direct contact with the RNA polymerase and secure it to the promoter.[2] [three] [4]
Eukaryotes [edit]
In eukaryotes, activators have a diversity of dissimilar target molecules that they can recruit in lodge to promote gene transcription.[1] [two] They tin recruit other transcription factors and cofactors that are needed in transcription initiation.[1] [2]
Activators can recruit molecules known every bit coactivators.[1] [2] These coactivator molecules can then perform functions necessary for beginning transcription in place of the activators themselves, such as chromatin modifications.[1] [2]
Deoxyribonucleic acid is much more than condensed in eukaryotes; thus, activators tend to recruit proteins that are able to restructure the chromatin and then the promoter is more than easily accessible by the transcription mechanism.[one] [2] Some proteins volition rearrange the layout of nucleosomes along the Dna in order to expose the promoter site (ATP-dependent chromatin remodeling complexes).[1] [2] Other proteins affect the bounden between histones and Dna via post-translational histone modifications, allowing the DNA tightly wrapped into nucleosomes to loosen.[one] [2]
All of these recruited molecules work together in order to ultimately recruit the RNA polymerase to the promoter site.[i] [2]
Release of RNA polymerase [edit]
Activators tin can promote gene transcription by signaling the RNA polymerase to move beyond the promoter and proceed along the Dna, initiating the first of transcription.[2] The RNA polymerase can sometimes interruption shortly after starting time transcription, and activators are required to release RNA polymerase from this "stalled" state.[ane] [2] Multiple mechanisms be for releasing these "stalled" RNA polymerases. Activators may act simply as a signal to trigger the continued move of the RNA polymerase.[2] If the DNA is likewise condensed to allow RNA polymerase to proceed transcription, activators may recruit proteins that can restructure the Dna so any blocks are removed.[i] [ii] Activators may also promote the recruitment of elongation factors, which are necessary for the RNA polymerase to continue transcription.[1] [2]
Regulation of activators [edit]
There are different means in which the action of activators themselves can exist regulated, in order to ensure that activators are stimulating gene transcription at advisable times and levels.[1] Activator activity can increase or subtract in response to environmental stimuli or other intracellular signals.[i]
Activation of activator proteins [edit]
Activators often must exist "turned on" before they tin can promote gene transcription.[2] [3] [4] The activity of activators is controlled by the power of the activator to bind to its regulatory site along the Deoxyribonucleic acid.[2] [iii] [4] The Deoxyribonucleic acid-binding domain of the activator has an active form and an inactive grade, which are controlled past the binding of molecules known as allosteric effectors to the allosteric site of the activator.[4]
Activators in their inactive grade are not bound to whatever allosteric effectors.[4] When inactive, the activator is unable to bind to its specific regulatory sequence in the Dna, and thus has no regulatory issue on the transcription of genes.[4]
When an allosteric effector binds to the allosteric site of an activator, a conformational change in the DNA-binding domain occurs, which allows the protein to bind to the DNA and increase gene transcription.[2] [four]
Mail-translational modifications [edit]
Some activators are able to undergo mail-translational modifications that have an result on their activity within a cell.[one] Processes such every bit phosphorylation, acetylation, and ubiquitination, amongst others, accept been seen to regulate the activity of activators.[1] Depending on the chemic grouping being added, every bit well as the nature of the activator itself, post-translational modifications can either increase or decrease the activity of an activator.[1] For example, acetylation has been seen to increase the activity of some activators through mechanisms such as increasing DNA-binding affinity.[1] On the other hand, ubiquitination decreases the activity of activators, as ubiquitin marks proteins for degradation after they have performed their corresponding functions.[1]
Synergy [edit]
In prokaryotes, a alone activator poly peptide is able to promote transcription.[ii] [three] In eukaryotes, usually more than 1 activator assembles at the bounden-site, forming a complex that acts to promote transcription.[1] [2] These activators bind cooperatively at the bounden-site, pregnant that the binding of one activator increases the affinity of the site to bind another activator (or in some cases some other transcriptional regulator) thus making it easier for multiple activators to bind at the site.[1] [2] In these cases, the activators interact with each other synergistically, meaning that the rate of transcription that is achieved from multiple activators working together is much higher than the condiment effects of the activators if they were working individually.[1] [2]
Examples [edit]
Regulation of maltose catabolism [edit]
The breakup of maltose in Escherichia coli is controlled by factor activation.[3] The genes that code for the enzymes responsible for maltose catabolism can just be transcribed in the presence of an activator.[3]
The activator that controls transcription of the maltose enzymes is "off" in the absence of maltose.[3] In its inactive form, the activator is unable to demark to DNA and promote transcription of the maltose genes.[iii] [4]
When maltose is present in the cell, it binds to the allosteric site of the activator protein, causing a conformational change in the Dna-binding domain of the activator.[iii] [4] This conformational alter "turns on" the activator past allowing it to demark to its specific regulatory Dna sequence.[iii] [four] Bounden of the activator to its regulatory site promotes RNA polymerase binding to the promoter and thus transcription, producing the enzymes that are needed to break downwardly the maltose that has entered the cell.[3]
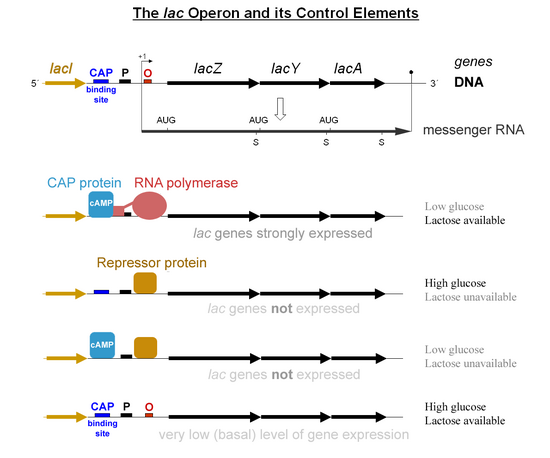
Regulation of the lac operon [edit]
The catabolite activator poly peptide (CAP), otherwise known as campsite receptor protein (CRP), activates transcription at the lac operon of the bacterium Escherichia coli.[5] Cyclic adenosine monophosphate (cAMP) is produced during glucose starvation; this molecule acts as an allosteric effector that binds to CAP and causes a conformational change that allows CAP to bind to a Dna site located adjacent to the lac promoter.[5] CAP and so makes a direct protein–protein interaction with RNA polymerase that recruits RNA polymerase to the lac promoter.[5]
See also [edit]
- Coactivator (genetics)
- Glossary of gene expression terms
- Operon
- Promoter (biology)
- Regulation of gene expression
- Repressor
- Transcription factor
- Bacterial transcription
- Eukaryotic transcription
References [edit]
- ^ a b c d due east f one thousand h i j yard 50 thou north o p q r s t u v w x y z aa ab air-conditioning ad ae af ag ah ai aj ak al am an Ma, Jun (2011). "Transcriptional activators and activation mechanisms". Protein & Jail cell. ii (11): 879–888. doi:10.1007/s13238-011-1101-seven. ISSN 1674-8018. PMC4712173. PMID 22180087 – via PubMed.
- ^ a b c d e f g h i j thou fifty m n o p q r southward t u five west x y z aa ab air conditioning ad ae af ag ah ai aj ak al am an ao ap aq ar as at au Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Morgan, David; Raff, Martin; Roberts, Keith; Walter, Peter (2015). Molecular Biology of the Cell (Sixth ed.). New York, NY: Garland Science. pp. 373–392. ISBN978-0-8153-4432-ii. OCLC 887605755.
- ^ a b c d east f thou h i j k l g northward o p q r due south t u 5 w x y z aa ab Madigan, Michael T; Bough, Kelly Southward; Buckley, Daniel H; Sattley, Matthew West; Stahl, David A (2018). Brock Biology of Microorganisms (Fifteenth ed.). NY, NY: Pearson. pp. 174–179. ISBN978-0-13-426192-8. OCLC 958205447.
- ^ a b c d e f g h i j yard fifty m n o p Griffiths, Anthony J.F.; Gelbart, William 1000.; Miller, Jeffrey H.; Lewontin, Richard C. (1999). "The Basics of Prokaryotic Transcriptional Regulation". Modern Genetic Analysis – via NCBI.
- ^ a b c Busby, Steve; Ebright, Richard H (1999-10-22). "Transcription activation past catabolite activator protein (CAP)". Journal of Molecular Biology. 293 (two): 199–213. doi:ten.1006/jmbi.1999.3161. ISSN 0022-2836. PMID 10550204.
Source: https://en.wikipedia.org/wiki/Activator_(genetics)
0 Response to "activator proteins bind to what type of dna sequence"
Post a Comment